Lab 2:
DNA Extraction
PRE-LAB CONCEPTS
Introduction
DNA Composition
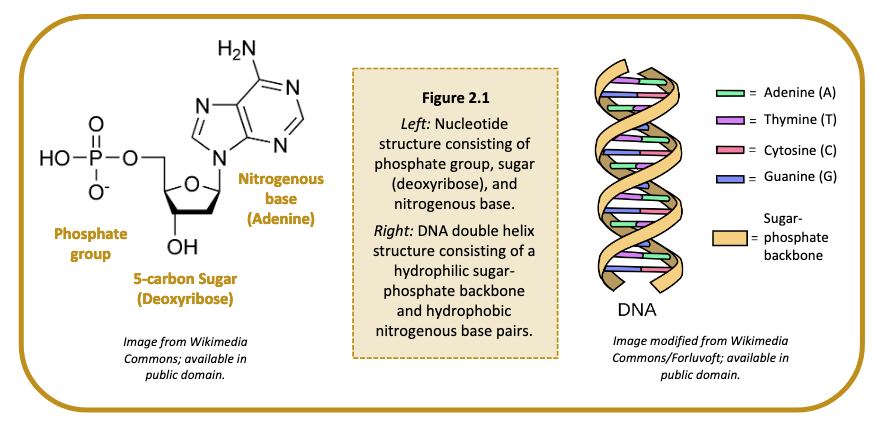
Deoxyribonucleic acid, or DNA, is a self-replicating molecule that encodes the genetic information for nearly all living organisms. DNA is comprised of monomers, called nucleotides. Each nucleotide consists of (i) a five-carbon sugar (deoxyribose), (ii) a phosphate group, and (iii) a nitrogenous base (Figure 2.1). Nucleotides are differentiated by their attached nitrogenous base: adenine (A), thymine (T), guanine (G) or cytosine (C). Adenine and guanine are classified as purines; cytosine and thymine are classified as pyrimidines. The nucleotides are linked together via phosphodiester bonds to form long strands of DNA, where the order of nucleotides along the sugar-phosphate backbone determines the DNA sequence.
DNA Structure
A single strand of DNA is asymmetrical and therefore has a specific method of combining with a second strand of DNA. The sugar and phosphate group are both hydrophilic and will readily come into contact with water molecules within a cell, whereas the nitrogenous bases are hydrophobic and will aggregate together. Two strands of DNA can satisfy their hydrophilic and hydrophobic constraints by orienting their nitrogenous bases to the inside and their hydrophilic sugars and phosphate groups to the outside. The nitrogenous bases are held together by hydrogen bonds, which maintain the shape of the DNA ladder. These base pairs are formed when a pyrimidine binds with a purine. Specifically, adenine forms a base pair only with thymine via two hydrogen bonds, and cytosine forms a base pair with only guanine via three hydrogen bonds (Figure 2.1). A skew and a twist of the strands of DNA lets in as little water as possible between the nitrogenous bases, forming the well-known double helix shape of the double stranded DNA (dsDNA) molecule.
Cellular DNA
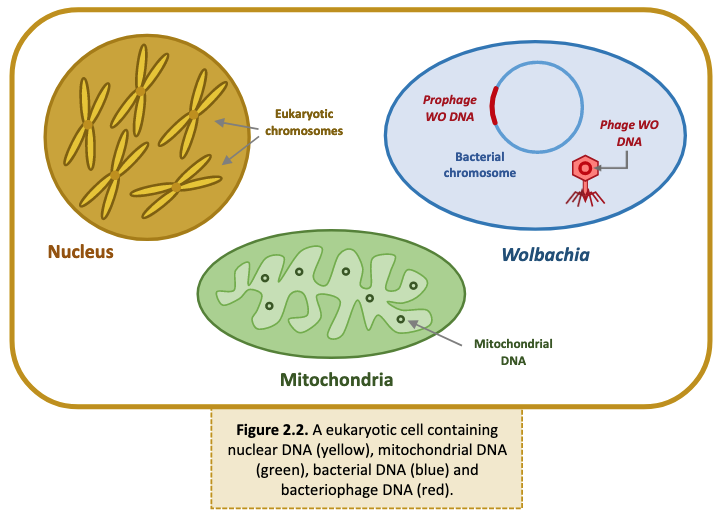
Arthropod cells may contain multiple types of DNA (Figure 2.2).
- Nuclear DNA: Eukaryotic DNA is coiled and condensed around nuclear proteins, called histones, to form chromosomes. Chromosomes are stored in the nucleus of each cell and encode the arthropod’s genetic information.
- Mitochondrial DNA: Mitochondria, the organelle responsible for cellular respiration and energy production, contains its own DNA. Mitochondrial DNA is circular and, like its encoding organelle, is passed down from mother to offspring.
- Bacterial DNA: Wolbachia and other bacterial endosymbionts contain a single, circular, supercoiled DNA molecule called the bacterial chromosome.
- Bacteriophage DNA: Phage WO, the bacteriophage that infects Wolbachia, carries its own genome. Phage WO is a temperate phage, meaning it can integrate its genome into the bacterial chromosome. When active, WO packages its DNA into phage capsids and lyses the Wolbachia cell to infect new hosts. Therefore, phage DNA can be found in both the Wolbachia chromosome and in phage particles.
In addition to the DNA shown above, an arthropod cell may also contain eukaryotic viruses and other intracellular bacteria (e.g., Rickettsia, Spiroplasma, Cardinium).
Wolbachia Localization
In most cases, whole body DNA extractions are sufficient to obtain Wolbachia DNA. If a specimen is particularly large and/or has a tough exoskeleton, abdominal dissections are recommended. A general rule of thumb is to dissect specimens that are larger than a grain of rice, such as a fruit fly, small ant, or mosquito. Because Wolbachia is maternally transmitted, it is commonly localized in the reproductive organs (ovaries and testes); however, increasing studies have also detected the symbiont in somatic, non-reproductive tissue. For the purpose of this lab activity, we recommend dissecting the reproductive areas. Make sure the arthropod is not still alive; freezing overnight and/or preserving in alcohol should be done prior to dissections and DNA extraction.
General guidelines for Wolbachia DNA extractions:
- Small arthropods: If the specimen is about the size of a grain of rice (such as a fruit fly or small ant ~2 mm), extract DNA from the entire body.
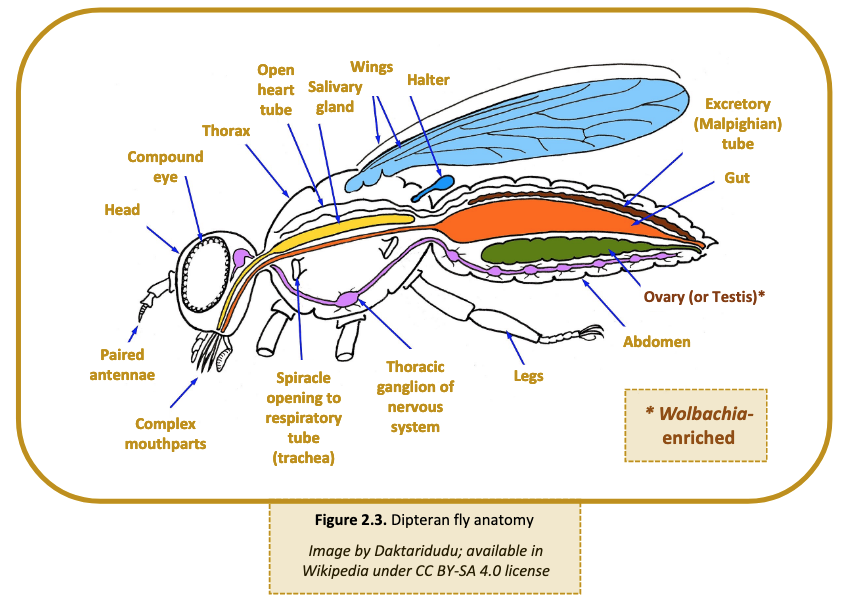
- Large arthropods: If the specimen is larger than ~2 mm, use a scalpel or razor blade to cut off the abdomen. In many cases, such as grasshoppers and cockroaches, the abdomen may still be larger than 2 mm. If so, use the scalpel to extract an internal portion of the abdomen most likely to contain reproductive tissue. The anatomy of a Dipteran fly is shown below for reference (Figure 2.3). Research the anatomy of each arthropod prior to dissections.
- Tough arthropods: If the specimen has a particularly tough exoskeleton, such as a tick, use a scalpel or razor blade to carefully cut open the abdomen and expose internal organs. This will ensure that the cell lysis buffer has access to reproductive tissues.
LAB PROTOCOL: DNA EXTRACTION
Based on feedback from teachers at all levels, silica-based spin kits deliver consistent and reliable results with less room for student error. Favorite kits include QIAGEN DNeasy Blood & Tissue Kit, NEB Monarch Genomic DNA Purification Kit, Promega Wizard SV Genomic DNA Purification Kit, and ThermoFisher GeneJET Genomic DNA Purification Kit. We recommend prioritizing a DNA extraction method that (i) has worked for you in the past; (ii) fits your budget; and/or (iii) is available to purchase in your area.
QIAGEN DNeasy Blood & Tissue Kit (Educator-favorite)
The protocol below has been enhanced with interactive Discovery Tools. Click on the highlighted text preceding the Discovery icon ![]() to view supporting media.
to view supporting media.
Sample Preparation
1. Collect four 1.5 ml microcentrifuge tubes and label them #1-4 ![]() . Note the contents of each tube in the chart below.
. Note the contents of each tube in the chart below.
| Tube # | Contents |
|---|---|
| 1 | |
| 2 | |
| 3 | Positive (+) Drosophila control |
| 4 | Negative (-) Drosophila control |
2. If preserved in alcohol, use tweezers to carefully transfer the first arthropod to a Petri dish. Rinse with water ![]() using either a transfer pipette or squirt bottle and blot dry excess liquid. Take 1-2 pictures for the The Wolbachia Project Database. If the arthropod is smaller than a grain of rice, skip to Step #4.
using either a transfer pipette or squirt bottle and blot dry excess liquid. Take 1-2 pictures for the The Wolbachia Project Database. If the arthropod is smaller than a grain of rice, skip to Step #4.
3. Remove the abdomen of the arthropod and cut off a small piece (roughly ~2 mm, or small enough to fit in the bottom of a microcentrifuge tube).
Remove as much preservative and water as possible. If the arthropod is large or has a thick/tough exoskeleton, dissect out the reproductive tissues. Arthropods with a thick exoskeleton should be cut into multiple pieces.
4. Place the specimen into a labeled 1.5 ml microcentrifuge tube and repeat for remaining arthropods.
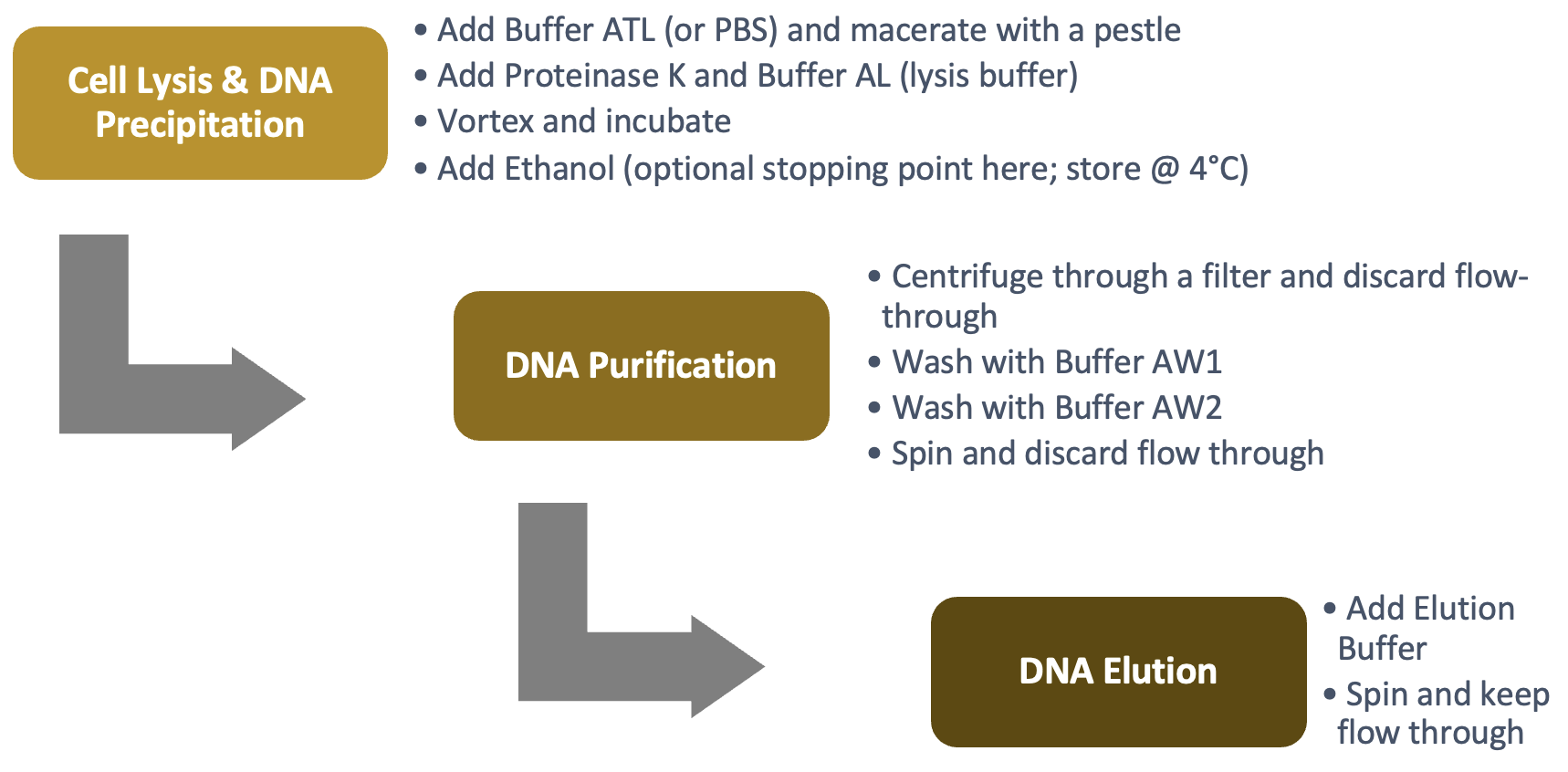
Cell Lysis & DNA Precipitation
5. Add 180 μl Buffer ATL ![]() to the first tube.
to the first tube.
Buffer ATL is a tissue lysis buffer.
6. Use a sterile pestle to grind the sample ![]() for 1 minute.
for 1 minute.
This is the most critical step of the entire protocol. Thorough grinding is necessary in order to obtain high DNA yield. Use muscle power and grind each sample thoroughly.
7. Using a new pestle for each tube; repeat Steps 5 and 6 with remaining samples.
8. Add 20 μl Proteinase K. ![]()
Proteinase K will destroy DNases that break down DNA.
9. Add 200 μl Buffer AL ![]() and immediately mix by vortexing for 10 seconds or pipetting up and down.
and immediately mix by vortexing for 10 seconds or pipetting up and down.
Buffer AL lyses open cells.
10. Incubate ![]() for at least 15 minutes @ 56 °C. Longer incubation times (i.e., 2-3 hours) are most effective for cellular lysis.
for at least 15 minutes @ 56 °C. Longer incubation times (i.e., 2-3 hours) are most effective for cellular lysis.
11. Centrifuge (spin) the tube for about 30 seconds to pellet debris ![]() as this could clog the filter of the spin column. Using a new tip for each sample, transfer the supernatant to a labeled 1.5 ml tube. Discard the old tube of cellular debris and repeat for other samples.
as this could clog the filter of the spin column. Using a new tip for each sample, transfer the supernatant to a labeled 1.5 ml tube. Discard the old tube of cellular debris and repeat for other samples.
Use caution to not disturb the pellet. Alternatively, pour the DNA-containing supernatant directly from each sample into the corresponding 1.5 ml tube.
12. Add 200 μl ethanol ![]() (96-100%) to each tube and mix by vortexing for 10 seconds or pipetting up and down.
(96-100%) to each tube and mix by vortexing for 10 seconds or pipetting up and down.
Ethanol precipitates DNA.
This is an optional STOPPING POINT. Store DNA in the refrigerator (4 °C) until next class period.
DNA Purification
13. Collect four DNeasy spin columns fitted with four 2.0 ml collection tubes and label the lids ![]() of the spin columns with your sample numbers.
of the spin columns with your sample numbers.
14. Pipet (or carefully pour) the liquid from Tube 1 of the above steps (including any precipitate) into the DNeasy Mini column #1. Using a new pipette tip for each transfer ![]() , repeat this process with the three other tubes. Make sure to keep the tube numbers consistent.
, repeat this process with the three other tubes. Make sure to keep the tube numbers consistent.
Throughout the protocol, be careful not to touch the filter or get liquid around the rim of the column.
15. Centrifuge all tubes for 1 minute ![]() at ≥6,000 x g (8,000 rpm). The DNA is now caught in the filter of the spin column. Carefully remove the spin columns from the collection tubes and discard the flow through (liquid) from the 2.0 ml collection tubes into the waste cup. Replace the spin columns back into their emptied collection tubes.
at ≥6,000 x g (8,000 rpm). The DNA is now caught in the filter of the spin column. Carefully remove the spin columns from the collection tubes and discard the flow through (liquid) from the 2.0 ml collection tubes into the waste cup. Replace the spin columns back into their emptied collection tubes.
If using a mini-centrifuge, you may not be able to fit all columns in the rotor. Spin 2-3 at once, ensuring that the centrifuge is properly balanced at all times. If your centrifuge does not have an adjustable speed, use the default speed throughout the protocol.
16. Add 500 μl of Buffer AW1 ![]() to each spin column.
to each spin column.
Buffer AW1 washes the DNA.
17. Centrifuge all tubes for 1 minute at ≥6,000 x g (8,000 rpm).
18. Again, discard the flow through waste from the 2.0 ml collection tubes into the waste cup and place the DNeasy Mini Spin Columns back into the same emptied 2.0 ml collection tubes.
19. Add 500 μl Buffer AW2 ![]() (a second wash buffer) to each of the four tubes and centrifuge for 3 minutes at 20,000 x g (14,000 rpm), or max speed of your centrifuge if it doesn’t go that high, to dry the DNeasy membrane.
(a second wash buffer) to each of the four tubes and centrifuge for 3 minutes at 20,000 x g (14,000 rpm), or max speed of your centrifuge if it doesn’t go that high, to dry the DNeasy membrane.
If the centrifuge you are using cannot attain this speed, allow the tube to air dry for 5 minutes. This will evaporate the ethanol. It is important to air dry the membrane so that ethanol does not interfere with PCR.
DNA Elution
20. Transfer the spin columns ![]() into 1.5 ml microcentrifuge tubes. Be sure to label the lids of each tube #1-4 and include your group number (or initials). These will contain your purified DNA samples.
into 1.5 ml microcentrifuge tubes. Be sure to label the lids of each tube #1-4 and include your group number (or initials). These will contain your purified DNA samples.
Carefully remove the DNeasy column so that it does not make contact with residual ethanol in the tube. If ethanol is still present on the membrane, empty the collection tube and spin again.
At this point, you will have two sets of lids. When you place them in the rotor, rotate the column such that its lid is not directly on top of the 1.5 ml tube lid. Keep lids down against the center of the rotor, not sticking out, and face them in the direction of motion. If a lid breaks off, transfer DNA to a new labeled 1.5 ml tube.
21. Pipet 100 μl of Buffer AE ![]() directly onto the spin column membrane. Incubate at room temperature for 1 minute.
directly onto the spin column membrane. Incubate at room temperature for 1 minute.
Buffer AE is is an elution buffer that rinses the DNA off the spin column filter and into the 1.5 ml tube. To incubate at room temperature, simply leave the tube sitting on the lab bench for 1 minute.
22. Centrifuge ![]() at ≥6,000 g or 8,000 rpm for 1 minute. The eluted DNA will be collected in the 1.5 ml tube.
at ≥6,000 g or 8,000 rpm for 1 minute. The eluted DNA will be collected in the 1.5 ml tube.
23. Discard the spin column and KEEP the labeled 1.5 ml tube. The DNA is now in the 1.5 ml microcentrifuge tube. ![]() Optional: Incubate the DNA for 1 hour @ 65 °C or overnight at room temperature.
Optional: Incubate the DNA for 1 hour @ 65 °C or overnight at room temperature.
24. Store the eluted DNA frozen at -20 °C until PCR.
(A condensed Bench Protocol is available on page 12 of the Student Guide below)
Shared Class Materials
- Incubator, heat block
 , or water bath, 56 °C
, or water bath, 56 °C - Vortex mixer

- Mini-centrifuge

- Float Rack *

- Metal tongs *
(* only needed if using a water bath)
Materials per Group
- 2 Arthropod specimens
- +/- Drosophila controls

- Gloves

- Tweezers
 / Scalpel
/ Scalpel - Petri dish

- Water
- Tranfer pipette
 or squirt bottle
or squirt bottle 
- 4 Microtube pestles

- 12- 1.5 ml tubes

- Qiagen DNeasy Kit
 aliquots
aliquots
• Buffer ATL (900 μl)
(900 μl)
• Proteinase K (100 μl)
(100 μl)
• Buffer AL (1.0 ml)
(1.0 ml)
• Buffer AW1 (2 x 1.25 ml)
(2 x 1.25 ml)
• Buffer AW2 (2 x 1.25 ml)
(2 x 1.25 ml)
• Buffer AE (500 μl)
(500 μl) - Ethanol
 (95-100%, 1.0 ml)
(95-100%, 1.0 ml) - 4 spin columns

- 4 collection tubes

- P200 and P1000 pipettes

- P200
 and P1000 tips
and P1000 tips 
- Waste cup
 for tips
for tips - Waste cup
 for liquids
for liquids - Sharpie

- Kimwipes

- Tube rack

This DNA extraction protocol is based on the insect adaptation of Qiagen’s DNeasy Blood & Tissue kit (Product # 69504).
Sample Preparation
Each specimen will be rinsed with water to remove alcohol preservative. If larger than a fruit fly, the abdomen will be dissected to obtain the reproductive tissues. If the specimen contains a particularly tough exoskeleton, abdominal tissue can be exposed using a scalpel or razor blade.
Cell Lysis & DNA Precipitation
Each specimen will be macerated in a cell lysis solution to break open the cellular and nuclear membranes. As a result, DNA is exposed to proteases, such as nucleases, in the host tissue. Therefore, the enzyme Proteinase K must be added to denature the proteins and keep the DNA intact. The solution is heated to ~ 56°C for enhanced lysis. Finally, ethanol will be added to bring the DNA out of solution.
DNA Purification
Once the proteases are destroyed and DNA is precipitated, the DNA must be purified. All cellular components, including DNA, will be placed into a spin column. Upon centrifugation, the material will pass through the filter, which attracts DNA but allows cellular debris to pass through. This will be followed by two wash steps with two separate buffers, AW1 and AW2.
DNA Elution
The activity will be completed by removing the DNA from the filter. This is called eluting the DNA and is done by adding the elution Buffer AE. DNA is more attracted to the elution buffer than the filter, and finally passes through the filter. Spinning the tube with the DNA embedded in the filter will pull the elution buffer through the matrix, thus collecting the DNA into the 1.5 ml tube.
ALTERNATIVE PROTOCOLS
FAQs